– Identification of enzyme modification sites to improve production rates of target compounds –
Researchers) IKEBE Jinzen, Researcher, KAMEDA Tomoshi, Senior Researcher, Computational Omics Research Team, Artificial Intelligence Research Center
- Identification of amino acid sites that control enzyme reactions by computer simulation
- Demonstration of increase of the production rate of target compounds with modified enzymes avoiding the formation of unwanted compounds
- Contribution to the reduction of labor required for preparation and evaluation of enzymes
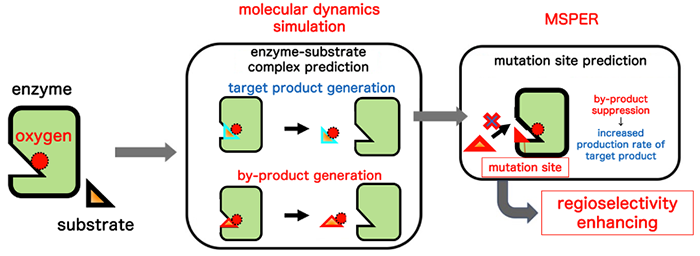
Principle of MSPER: A noble method to increase production rate of a target compound in enzyme reactions using computer-assisted enzyme modification
Bio-manufacturing uses enzyme reactions to produce compounds that serve as materials such as pharmaceuticals, foods, fibers, and plastics. It is conducted using aqueous solvents under normal temperature and pressure conditions, as compared to conventional chemical reactions using petroleum-derived organic solvents under high temperature and pressure conditions. For this reason, bio-manufacturing draws attention as a next-generation technology that can be to reduce utility costs and environmental impacts such as waste liquid treatment. Further improvement of enzyme function is important for the industrial use of enzymes.
Enzymes form specific complex structures with their substrate for the enzymatic reaction. Depending on the types of enzymes and substrate, several different complexes may be formed causing multiple product formations, and the formation of by-products reduces the production rate of the target compound. To address this issue, it is necessary to find and modify the amino acid sites involved in the enzyme function from among the hundreds to thousands of amino acids that constitute the enzyme. The evolutionary molecular engineering method and various rational design methods have been devised as methods for achieving this. The evolutionary molecular engineering method repeatedly produces and conducts verification tests of various modified enzymes, so enormous time, labor, and costs are needed to obtain the optimal modified enzyme. In contrast, rational design can save the labor for verification tests by predicting the amino sites that modify enzyme functions from information such as the three-dimensional structures of enzymes and substrates. However, methods specialized in the suppression of by-products had yet to be developed.
Researchers in AIST has developed a computational method for predicting the amino acid sites that control enzyme reactions (Mutation Site Prediction method for Enhancing the Regioselectivity of substrate reaction sites, MSPER) in collaboration with KNC Laboratories Co., Ltd. This method was applied to modify the enzyme cytochrome P450 (P450) used in the production of perfume raw materials. The production rate of the target compound was increased by up to 6.4 times compared to the unmodified enzyme.
MSPER was developed that suppresses the production of by-products by predicting and modifying amino acid sites involved in by-product production from the structural information obtained from simulation analysis to reproduce multiple enzyme-substrate complexes generated in enzyme reactions. This method enables to narrow down the enzyme areas to be modified, which achieves labor savings by reducing the number of evaluation tests for functional verification to 1/170 to 1/1000 that of conventional random mutation methods.