Update(MM/DD/YYYY):03/10/2023
Development of Technology to Quickly Determine the Health Status of Mice Using Gut Microbiota
– Use of machine learning to analyze the fluorescence patterns of polymers contacted with bacteria –
Researchers) TOMITA Shunsuke, Senior Researcher, KOJIMA Naoshi, Senior Researcher, ISHIHARA Sayaka, Technical Staff, KURITA Ryoji, Group Leader, Nano-biodevice Research Group, Health and Medical Research Institute, KUSADA Hiroyuki, Researcher, TAMAKI Hideyuki, Group Leader, Microbial and Genetic Resources Research Group, Bioproduction Research Institute, MIYAZAKI Koyomi, Deputy Director General, Department of Life Science and Biotechnology
Points
- Development of a group of polymers that can convert bacterial surface characteristics into blue fluorescence simply by mixing with gut microbiota
- Analysis of fluorescence intensity patterns by machine learning enabled accurate discrimination between the gut microbiota of healthy mice from those of insomniac mice
- This technology holds promise for quick, easy, and inexpensive assessing health status without harming patients
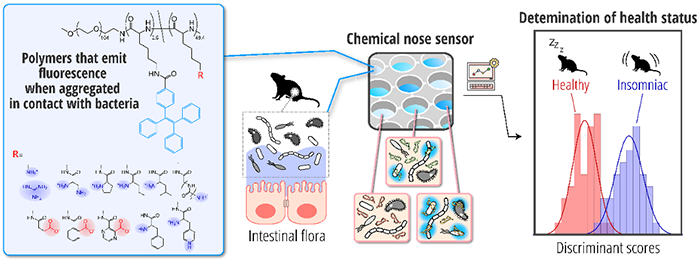
Analysis of the gut microbiota of mice by the developed chemical nose sensor
Background
The human intestine is home to more than 1,000 species of diverse bacteria, which are collectively referred to as “gut microbiome.” Gut microbiotas interact with the human host and are deeply involved in health maintenance and disease development. Therefore, understanding and controlling the characteristics of the bacterial composition of the gut microbiota is of great importance from the perspective of preventive medicine.
A standard method for analyzing gut microbiota is 16S ribosomal RNA (rRNA) amplicon sequencing analysis, which enables to detect a large number of bacteria that constitute the gut microbiota by sequencing a PCR-amplified marker gene (16S rRNA gene). However, this analysis method requires expensive next-generation sequencers, specialized knowledge, and much labor for data acquisition and analysis. Therefore, it is currently unable to meet the needs of industrial and medical settings that require quick assessment of gut microbiome status with limited equipment.
Summary
Researchers in AIST developed a novel analytical technology that can determine the characteristics of the bacterial composition of gut microbiota with high accuracy, using polymers that emit blue fluorescence when in contact with bacteria and machine learning to screen the characteristics of the fluorescence intensity patterns.
This technology uses a bioanalytical method called a chemical nose. The chemical nose developed consists of 12 types of polymers with fluorophores that emit light when aggregating. By mixing these polymers with intestinal bacteria, various fluorescent signals can be detected, and the bacteria can be characterized based on those patterns. Using the developed chemical nose, the researchers succeeded in determining the health status of mice with a high degree of accuracy by comparative analysis of gut microbiome samples collected from healthy and insomniac mice. This technology enabled to characterize the state of gut microbiota from a different perspective than the standard gut microbiome analysis method (e.g., 16S rRNA gene amplicon sequencing analysis), and has the advantages of being faster, easier, and less expensive than amplicon sequencing analysis. In the future, it is expected to be applied as a diagnostic technique for health care, using human gut microbiome samples as specimens.