Takema Fukatsu (Prime Senior Researcher and Leader of Symbiotic Evolution and Biological Functions Research Group), Ryuichi Koga (Senior Researcher), and others of the Bioproduction Research Institute (Director: Yoichi Kamagata) of the National Institute of Advanced Industrial Science and Technology (AIST; President: Ryoji Chubachi) and Noriyuki Sato (Professor) and others of Okinawa Institute of Science and Technology Graduate University (OIST; President: Jonathan Dorfan), in collaboration with the University of Montana (USA), the Open University of Japan, and others, have discovered that two types of bacteria with extremely reduced genomes endocellularly reside in a nested manner within the symbiotic organ of mealybugs that are known as agricultural pests, and that more than 20 different genes laterally transferred from various bacteria to the insect’s genome in the past are expressed in the symbiotic organ. The researchers also found that these symbiont genomes and laterally transferred bacterial genes together form mutually complementary metabolic pathways essential for the symbiotic relationship in a mosaic manner, which are involved in synthesis of amino acids, vitamins, and cell walls. Such a complexity of the symbiotic system is beyond what has been conventionally believed.
These results have been published in an American scientific journal, Cell, on June 21, 2013 (JST). (DOI:10.1016/j.cell.2013.05.040)
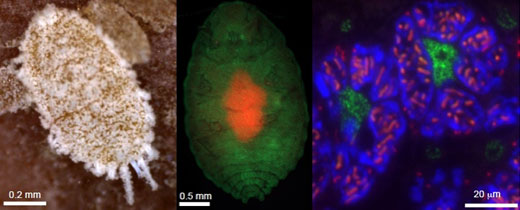 |
|
(Left) The citrus mealybug (Planococcus citri). (Center) In the abdomen, an oval symbiotic organ, called the bacteriome consisting of many bacteriocytes (red), is located. (Right) The cytoplasm of each bacteriocyte harbors β-symbiotic bacterial cells amorphous in shape (blue), and each β-symbiont cell further contains rod-shaped γ-symbiotic bacterial cells (red) in a nested manner, while host’s cell nuclei (green) encode laterally transferred genes derived from various bacteria. |
The most fundamental classification of the great variety of organisms living on earth is the distinction between prokaryotes and eukaryotes. Elucidating the processes through which complex eukaryotic cells appeared from simpler prokaryotic cells is pivotal for understanding the diversity and evolution of life. The endosymbiotic theory has been established as the basic explanation for the origin of eukaryotes. A wide variety of endosymbiotic relationships have been found in insects, plants, fungi, protists, etc. In all these cases, eukaryotic cells incorporate prokaryotic cells or other eukaryotic cells as endosymbionts. For many years, no examples had been known of endosymbiosis in which a prokaryotic cell incorporated another prokaryotic cell, a phenomenon that is potentially relevant to our understanding of the evolutionary process of eukaryotic cells.
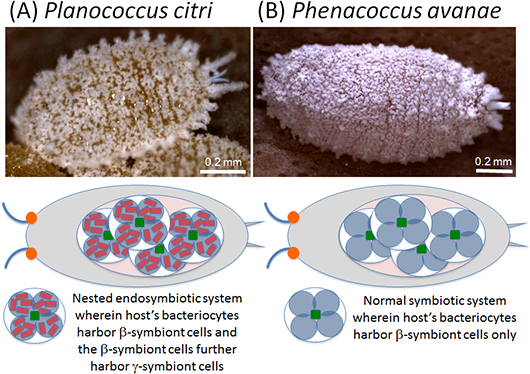
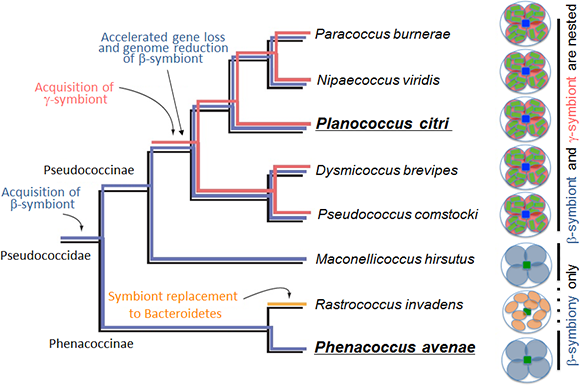
However, it was discovered that the citrus mealybug (Planococcus citri), known as an agricultural pest, harbors β-proteobacterial symbiont Tremblaya (β-symbiont) within specialized cells called bacteriocytes that constitute the bacteriome, a symbiotic organ, in the abdomen, and the Tremblaya cells further harbor γ-proteobacterial symbiont Moranella (γ-symbiont) endocellularly, thereby forming a nested symbiotic system. Subsequently, genome sequences the β-symbiont and the γ-symbiont of Planococcus citri were determined. Comparative studies on the endosymbiotic systems of various mealybug species revealed that members of the subfamily Pseudococcinae, including Planococcus citri, possess the nested endosymbiotic system consisting of the γ-symbiont cells within the β-symbiont cells, whereas members of the subfamily Phenacoccinae, including the oat mealybug (Phenacoccus avenae), possess a normal endosymbiotic system consisting of the only β-symbiont cells within the bacteriocytes. These results suggested the possibility that the endosymbiotic system of the Phenacoccinae may represent an ancestral state before acquiring the γ-symbiont (Figs. 1, 2).
 |
|
Figure 1 : Schematic diagrams of the endosymbiotic system in the citrus mealybug (Planococcus citri) (A) and the oat mealybug (Phenacoccus avenae) (B) |
 |
|
Figure 2 : Diversity and evolutionary processes of the endosymbiotic systems in the mealybug family Pseudococcidae |
The Japanese research group headed by AIST started a research project in collaboration with the American research group led by the University of Montana to perform comprehensive gene expression analysis and draft genome sequencing of the symbiotic organ of Planococcus citri (Fig. 1A). The Japanese group also determined the complete genome sequence of the β-symbiont of Phenacoccus avenae in collaboration with a research team from OIST (Fig. 1B). In this way, the international collaborative project aimed at elucidating the evolutionary processes of the endosymbiotic system consisting of prokaryotic cells with other prokaryotic cells among mealybugs at the genomic level.
This research was partly supported by the Bio-oriented Technology Research Advancement Institution of the National Agriculture and Food Research Organization, and also by a scientific research grant from the Ministry of Education, Culture, Sports, Science and Technology.
The β-symbiont genome of Phenacoccus avenae harbored 175 protein-coding genes. Considering that the β-symbiont genome of Planococcus citri contains 121 protein-coding genes, it is conjectured that the β-symbiont has lost at least 50 genes since the acquisition of the γ-symbiont (Fig. 2).
Whereas the β-symbiont of Phenacoccus avenae possessed most of the genes involved in translation (Fig. 3B, blue), the β-symbiont of Planococcus citri has lost nearly all translation-related genes, which were mostly complemented by the corresponding genes of the γ-symbiont (Fig. 3A, orange). Hence, It is conjectured that the β-symbiont has lost the translation-related genes after the acquisition of the γ-symbiont.
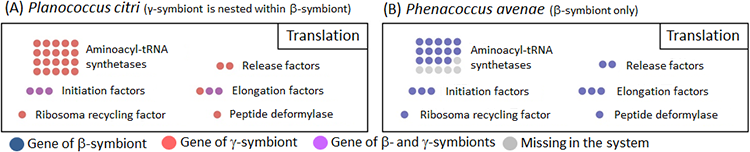 |
Figure 3 : Configuration of translation-related genes in the endosymbiotic system of Planococcus citri (A) and Phenacoccus avenae (B)
Dot color indicates source of the gene or presence/absence of the gene (see the keys). The number of dots indicates the number of genes belonging to the category. For example, there are 20 genes that encode aminoacyl-tRNA synthetases, corresponding to 20 amino acids constituting proteins. |
Since mealybugs feed only on plant sap, which contains little protein, throughout their life, they require a supply of ten essential amino acids from their symbiotic bacteria. In the β-symbiont genome of Phenacoccus avenae, the biosynthetic pathways for essential amino acids were complete only for tryptophan and arginine, while the biosynthetic pathways for the other eight essential amino acids were incomplete (Fig. 4B). In Planococcus citri, similarly, tryptophan and arginine were the only two essential amino acids for which all biosynthetic pathway genes are available, but, notably, genes of the β-symbiont (Fig. 4A, orange) and genes of the γ-symbiont (Fig. 4A, blue) together constituted complete biosynthetic pathways, fitting together like a mosaic. For the other eight essential amino acids, genes of the β-symbiont and genes of the γ-symbiont likewise fitted together like a mosaic, but the biosynthetic pathways were incomplete (Fig. 4A).
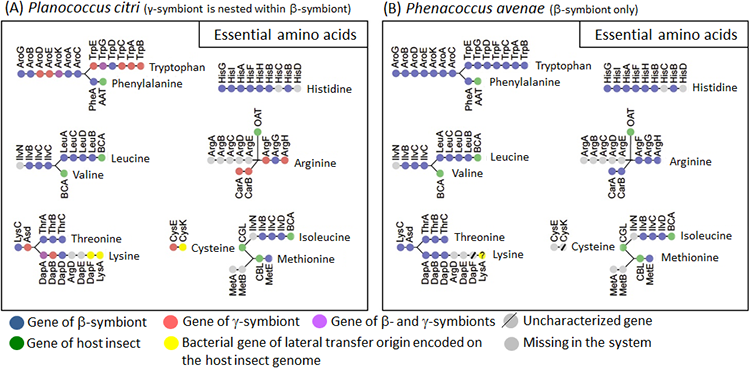 |
Figure 4 : Configuration of essential amino acid synthesis-related genes in the endosymbiotic system of Planococcus citri (A) and Phenacoccus avenae (B)
Dots represent the genes involved in synthesis of amino acids (gene names are indicated above or below the dots), and are arranged in the order found in the amino acid synthetic pathways. Dot color indicates source of the gene or presence/absence of the gene (see the keys). |
In an attempt to identify candidate genes that may be filling the gaps in the biosynthetic pathways for the essential amino acids constituted by the symbiont genes, comprehensive transcriptomic analyses were performed for the host insect genes expressed in the symbiotic organ and the whole body of Planococcus citri. The results clarified that, for some of the missing genes, host insect genes with similar functions are expressed in the symbiotic organ and appear to complement them (Fig. 4A, green). Meanwhile, it was also found that three of the missing genes are complemented not by host insect genes, but by genes derived from bacteria other than the β-symbiont and the γ-symbiont (Fig. 4A, yellow).
Similar results were obtained concerning the genes involved in the biosynthetic pathways for B vitamins, which are also important nutrients provided by the symbiotic bacteria (Fig. 5). Of the four genes of the riboflavin (vitamin B2) biosynthetic pathway in Planococcus citri, two were γ-symbiont genes (Fig. 5A, riboflavin/orange), but the other two were derived from bacteria other than the β-symbiont and the γ-symbiont (Fig. 5A, riboflavin/yellow). Similarly, in the biosynthetic pathway for biotin (vitamin B7), in addition to six γ-symbiont genes (Fig. 5A, biotin/orange), three final step genes (Fig. 5A, biotin/yellow) were derived from some other bacteria.
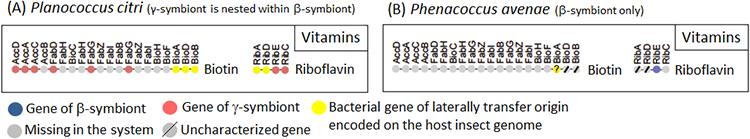 |
Figure 5 : Configuration of B vitamin (VB) synthesis-related genes in the endosymbiotic system of Planococcus citri (A) and Phenacoccus avenae (B)
Dots represent the genes involved in synthesis of B vitamins (gene names are indicated above the dots), and are arranged in the order found in the B vitamin synthetic pathways. Dot color indicates the source of the gene (see the keys). |
Even more extreme results were found for genes involved in cell wall biosynthesis (Fig. 6). In the symbiotic organ of Planococcus citri, only two genes involved in the biosynthesis of peptidoglycan, a major structural component of bacterial cell wall, were γ-symbiont genes (Fig. 6, orange), while all the other genes were derived from bacteria other than the β-symbiont and the γ-symbiont (Fig. 6, yellow).
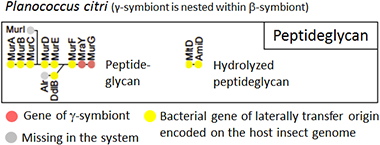 |
Figure 6 : Configuration of cell wall synthesis-related genes in the endosymbiotic system of Planococcus citri
Dots represent the genes involved in synthesis of cell walls (gene names are written above or below dots), and are arranged in the order found in the peptidoglycan synthetic pathway. Dot color indicates source of the gene or presence/absence of the gene (see the keys). |
Draft sequencing of the genome of Planococcus citri confirmed that these genes derived from some bacteria other than the β-symbiont and the γ-symbiont are all encoded in the host insect’s nuclear genome. Hence, mealybugs like Planococcus citri not only harbor two types of symbiotic bacteria nested in their cells, but also possess various metabolism-related genes that were acquired via lateral transfer from a wide variety of bacteria. These findings indicate that the mealybugs possess a symbiotic system far more complex than conventionally believed, in that the host insects are able to survive owing to functional metabolic pathways consisting of genes derived from two symbiotic bacteria and the laterally transferred genes.
Molecular phylogenetic analyses revealed that many of the laterally transferred bacterial genes are closely related to genes of such non-essential symbiotic bacteria widely found in insects as Wolbachia, Rickettsia, Sodalis, Arsenophonus, and Cardinium. Currently, however, Planococcus citri is not infected with these symbiotic bacteria. It is conjectured that perhaps an ancestor of Planococcus citri and allied mealybugs was infected with these symbiotic bacteria in the past, but the infections have been lost after transferring some genes to the host’s nuclear genome (Fig. 7).
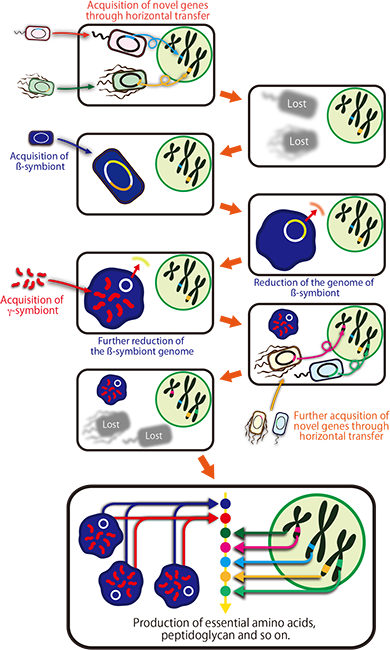 |
|
Figure 7 : Schematic diagram of the evolutionary processes underlying the endosymbiotic system of Planococcus citri |
These findings shed new light on the following fundamental questions: how should individuals, cells, and genomes of organisms be defined? How were these constructed? How have they evolved? The findings provide intriguing insights into the endosymbiotic theory, for example, which says that perhaps the greater part of symbiotic bacteria died out after just a portion of their genes were laterally transferred to the host’s nuclear genome, with just a part of them remaining as intracellular organelles like mitochondria and chloroplasts; that perhaps such laterally transferred genes also contributed to the evolution of eukaryotic cells; and that perhaps the earliest eukaryotic cells occurred as a chimera of multiple bacteria.
The researchers will conduct functional analysis on gene products such as proteins that are encoded by the host insect genes, the β-symbiont genes, the γ-symbiont genes, and laterally transferred genes that make up the endosymbiotic system of the mealybugs, thereby understanding how this complex symbiotic system is constructed and operating.
In addition, the researchers plan to explore endosymbiotic systems in various organisms to elucidate whether such a phenomenon is unique to the mealybugs or can also be found in other organisms, thereby pursuing the relationship between symbiotic evolution and lateral gene transfer.