Kentaro Miyazaki (Leader) and coworkers, Enzyme Exploration Research Group, the Bioproduction Research Institute (Director: Yoichi Kamagata) of the National Institute of Advanced Industrial Science and Technology (AIST; President: Tamotsu Nomakuchi), have discovered that ribosomes that have “translation” functions, synthesizing protein in living bodies based on genetic information, have a function of inhibiting the activity of an RNA-degrading enzyme, namely a ribonuclease (RNase).
Ribosomes play a major role in the “translation” of genetic information “transcribed” from DNA to RNA to protein. Meanwhile, RNase T2 is an RNA-degrading enzyme, which is involved in the inhibition of extracellular RNA invasion, incorporation of nutrition, etc. In E. coli (Escherichia coli), RNase T2 is present in the periplasmic layer located outside the intracellular membrane, and thus is isolated from intracellular RNA. However, RNase T2 enters cells and self-RNA can be degraded during the stationary phase or under stress. Accordingly, cells need a mechanism for protecting RNA from RNase T2, but the mechanism has been unknown.
It has been known that when RNase T2 is purified from E. coli, RNase T2 is isolated in an inactive form combined with a ribosome. However, the physiological significance of the formation of such a complex, its relation with translation functions, the mode of interaction, etc. have remained unknown. Through mutation analysis of 16S rRNA, the researchers have discovered that an interaction and/or inhibition-determining site is present in 16S rRNA in a 30S ribosomal subunit and intracellular self-RNA degradation is prevented by being combined with RNase T2.
These results will be published online in Nature Communications on November 23, 2011 (Japan time).
 |
|
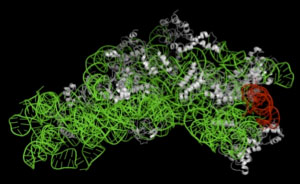
Figure 1 : Three-dimensional structure of 30S ribosomal subunit |
The green portion indicates 16S rRNA, the red portion indicates helix41 region in 16S rRNA interacting with RNase T2,
and the white portions indicate ribosomal protein. |
Ribosomes are cytoplasmic organelles existing in all organisms and have an important biological function, “translation.” Although ribosomal functions are common among many organisms, bacterial and human ribosomes have different structures. Hence, bacterial ribosome-selective inhibitors are potential anti-infective agents with low toxicity to human beings. Therefore, the development of such therapeutic agents has been expected, but it requires detailed analysis of ribosomal functions.
AIST has conducted various research concerning microorganisms such as searching for and using novel microorganisms and useful enzymes contained in them. It is currently elucidating and using cell functions through analysis and alteration of ribosomes using E. coli as a platform.
When E. coli RNase T2 is purified, it is isolated in a form combined with a ribosome that is thought to be unrelated to the physiological role of RNase T2. However, the physiological significance of interaction that causes binding of ribosomes with RNase T2 and the detailed molecular mechanism such as a mode for inhibition of RNase T2 have remained unknown. The researchers have elucidated the actual state of “RNase T2-ribosome interaction” that had long remained unsolved, as part of their analysis of ribosomal functions.
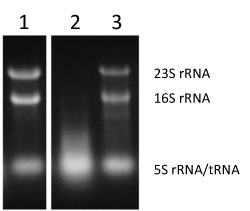
To elucidate the mechanism and physiological significance of the formation of RNase T2-ribosome complex, E. coli ribosome mutants were analyzed. First, E. coli 16S rRNA was substituted with a heterologous organism-derived 16S rRNA gene. Since 16S rRNA forms the central skeleton of the 30S ribosomal subunit, this mutation is thought to affect the entire ribosome. In addition, 16S rRNA is an essential gene, and so a drastic mutation can lead to a decrease in functions or cell death. 16S rRNA genes were cloned from various microorganisms and were then each substituted with the E. coli 16S rRNA gene. It was demonstrated that E. coli can grow even if the E. coli 16S rRNA gene is substituted with 16S rRNA derived from a microorganism evolutionarily distant from E. coli, such that the substituted gene has only about 80% sequence homology with the E. coli 16S rRNA gene. When the growth of these E. coli mutant strains was minutely observed, it was revealed that the strains tended to die during the stationary phase. Also, when intracellular RNA was extracted during the stationary phase, RNA degradation was confirmed in E. coli (KT103/Rpi) containing the mutation at a site for inhibiting RNase T2 (lane 2 in Fig. 2).
 |
|
Figure 2 : Evaluation of activity of inhibiting RNase T2 using intracellular RNA degradation as an index |
Lane 1 (KT103/Eco), lane 2 (KT103/Rpi), and lane 3 (KT103 rna-/Rpi).
RNA degradation (lack of bands indicating 23S rRNA and 16S rRNA) in lane 2 (KT103/Rpi). |
Next, to determine the region involved in the inhibition of RNase T2 activity, mosaic 16S rRNAs were synthesized (Fig. 3) from 16S rRNAs of E. coli and a heterologous organism, and the inhibition of RNase T2 activity was examined. Mosaic 16S rRNA prepared by substitution of a region referred to as helix41 (secondary structure formation region denoted in red in Fig. 1) in heterologous organism-derived 16S rRNA with E. coli 16S rRNA exhibited inhibiting activity and it has been revealed that the helix41 region plays a critical role in the inhibition of RNase T2 activity. It was further revealed by ex vivo binding experiments and inhibition experiments that interaction via helix41 is specific to RNase T2 and the inhibition of activity is induced by direct binding.
 |
|
Figure 3 : Synthesis of mosaic 16S rRNA gene derived from E. coli and heterologous organism |
|
1: E. coli 16S rRNA gene, 2: heterologous organism-derived 16S rRNA gene, 3: mosaic 16S rRNA gene from 1 and 2 |
RNase T2 is a protein which plays an important role in preventing foreign RNA from invading cells, but has a risk of entering cells and degrading intracellular self-RNA if the intracellular membrane is loosened or injured during the stationary phase or under stress conditions. Thus, it is thought that there is a mechanism by which ribosomes abundantly existing in cells have inhibitory activity, so as to protect self-RNA. Both RNase T2 and ribosome are biomolecules of ancient origin. RNase T2 and helix41 are thought to have co-evolved in a toxin-antitoxin relationship. It is considered that such non-translation function has long been imprinted in ribosomes, in addition to the functions as a translation apparatus.
Through this discovery of a new role of ribosomes which have conventionally been regarded as a translation apparatus for protein synthesis based on genetic information, it may be possible to identify other hidden important physiological functions in ribosomes. The researchers will continue to analyze ribosomal functions, elucidate the presence or absence of such unknown functions, and evaluate the potential application of ribosomes to anti-infective agents with low toxicity.