Senior researcher, P.K.R. Kumar et al. of National Institute of Advanced Industrial Science and Technology (hereafter referred to as AIST) 【president, Hiroyuki Yoshikawa】, Institute for Biological Resources and Functions【Director, Masahiro Iwakura】, in collaboration with National Institute of Agro biological Sciences (hereafter referred to as NIAS) 【president, Masaki Iwabuchi】, proposed a switch-on mechanism based on crystal structure of the Bacillus subtilis HutP anti-termination complex (a complex formed in the process of anti-termination, one of transcription control mechanism).
Genes responsible for L-histidine degradation and utilization, as carbon and nitrogen sources under nutrient-limited conditions, are located within the hut operon in Bacillus subtilis. The hut operon consists of five structural genes, hutH, hutU, hutI, hutG and hutM, and a positive regulatory gene hutP. The hutP gene is located just downstream of the promoter, whereas the structural genes are further downstream (Fig. 1). An overlapping nucleotide sequence, located between the hutP gene and the structural genes, is predicted to form an antitermination/terminator (stem-loop) structure depending upon the availability of L-histidine and Mg2+ ions. The transcription of the hut structural genes are regulated by HutP.
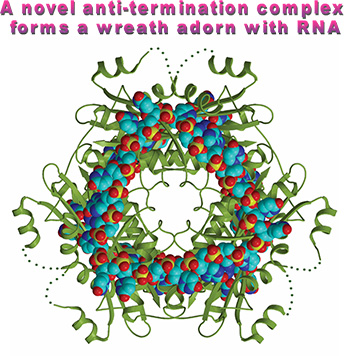
The quarternary structure (HutP-L-histidien-Mg2+-RNA) shows how HutP specifically recognizes the conserved sequences within the hut mRNA (Fig. 2), and reveals the unexpected direct role of the Mg2+ ion for mediating the L-histidine-dependent structural rearrangement in the protein. To unravel these structural changes, we have solved two additional crystal structures (uncomplexed HutP and HutP-L-hisitidine-Mg2+) and found that the Mg2+ ion coordinates with the L-histidine to facilitate an appropriate structural rearrangement before its recognition of the cognate RNA (Fig. 3). Once HutP has undergone this structural rearrangement, it binds specifically to the RNA sequence within the terminator region and wraps the RNA around the protein; by this, the RNA structure may reorganize and destabilize the terminator structure.
The present study appeared on the March 10th 2005 issue of Nature.
*Kumarevel, T., Mizuno, H. and Kumar, P.K.R. (2005) Structural basis of HutP-mediated anti-termination and roles of the Mg
2+ ion and L-histidine ligand. Nature
434, 183-191 (2005)
 |
|
Figure 1. Schematic representation of the hut operon showing the arrangement of the hutP, terminator/antiterminator region (stem-loop), and structural genes. |
Although bacteria frequently use attenuation/antitermination pathways to regulate their operons, the regulation and activation mechanisms of proteins that control alternative structures by RNA-binding protein were largely unknown. At present, the attenuation/antitermination protein-RNA complex structures are available for only two proteins, including the full-length trp RNA-binding attenuation protein (TRAP) and the amino-terminal peptide fragment of LicT. However, the structure in the absence of ligand (L-tryptophan) for the full-length TRAP is not available to reveal insights into any structural rearrangement that might occur on binding to their respective ligands. We sought to unravel the mechanism how these proteins are regulated for sensing the target sequence within the mRNA. For this purpose, we used HutP, a protein that regulates transcription by an antitermination, and clarified how the protein conformational changes recognize target sequence within the hut mRNA using crystallography studies.
Senior Researcher, P.K.R. Kumar et al. of Institute for Biological Resources and Functions, AIST have been studying RNA-protein interactions essential for gene regulations in various organisms and also found novel RNA motifs that binds to therapeutically important proteins (which are not known to bind to RNA) to generate novel reagents for diagnosis. Overal l understanding of RNA-protein specific-interactions, crystallographic studies are indispensable. For this purpose, collaboration with Hiroshi Mizuno, team leader of Department of Biochemistry, NIAS has been extended, who is now a research fellow of NEC Soft Ltd. This research is partly supported by Science and Technology Promoting Project, “ORCS” of the Ministry of Education, Science, Sports and Culture.
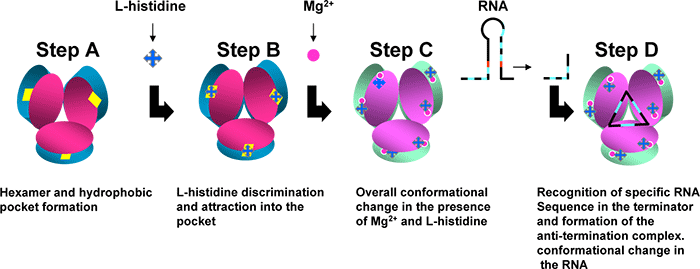
The present X-ray crystal structure analyses on HutP complexes showed the intermediate state structures before binding to the terminator/anti-termination RNA: Fig. 3 anti-termination complex, Fig. 3 HutP- L-histidine-Mg2+ ion, Fig. 3 HutP- L-histidine, Fig. 3 uncomplexed HutP. In these structures, HutP forms a hexamer Fig. 3, structure of HutP changes in the presence of L-histidine and Mg2+ ion Fig. 3, structurally reorganized HutP binds specifically to target RNA (a repeat sequence of U(uracil), A(adenine and G(guanine)) Fig. 3, the bound RNA forms a novel triangle fold on the surface of the HutP hexamer Fig. 3. In short, these studies clearly show that HutP initially undergoes structural rearrangement, brought by L-histidine and Mg2+ ion, for binding to target RNA sequence. This appears to lead structural changes in RNA structure (terminator structure) causing switch-on of the transcription of the downstream hut structural genes.
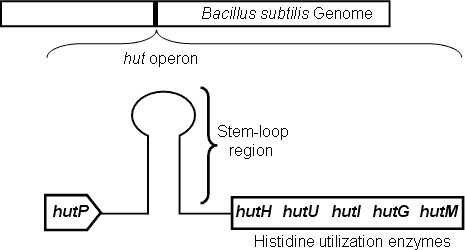 |
Figure 2. Hexameric HutP quarternary complex, viewed along the three-fold axis.
RNA and protein are shown in cpk and ribbon models, respectively. |
 |
|
Figure 3. A schematic model proposed for HutP antiterminator complex formation. |
In order to understand the diversity of proteins, which regulates alternative structures of mRNA, that regulate different operons, it is important to identify this class of proteins in whole genome and to analyze the mechanism of their regulation.