- Giving Impacts to Both Basic and Application Aspects of Protein Research as Well as to Studies on Origin of Life -
The Institute for Biological Resources and Functions (IBRF) and the Age Dimension Research Center (ADRC) of the National Institute of Advanced Industrial Science and Technology (AIST), an independent administrative institution, have designed and synthesized a new non-cyclic peptide consisting of 10 amino acid residues, based on an original algorithm developed at the AIST, and demonstrated that the peptide formed a stable 3D structure in aqueous solution and underwent reversible and cooperative denaturation/renaturation as a function of temperature. (See Fig. 1).
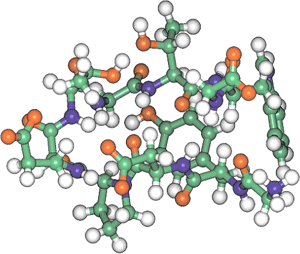 |
|
Fig. 1. 3D structure of newly designed peptide “chignolin”
PDB accession no.: 1UAO
|
It is a great mystery how proteins closely connected to the origin of life were created, and how they evolved to highly sophisticated molecules as we see today. The newly designed and synthesized peptide with molecular weight (MW) about 1,000 is much smaller than naturally occurring proteins with MW 5.000~150,000. (Fig. 2) However, in view of two essential requirements for protein functioning: specific 3D structures and cooperative structural transition, the synthesized peptide may be regarded as “a smallest protein”.
It has been generally conceived that for a protein keeping stable 3D structures, it has to possess at least 30~50 amino acid residues. The synthesized peptide pushes the lower limit widely downward, requesting to revise the understanding on the minimal structural unit of protein. The present study is expected to promote the progress in the studies of stabilization mechanism, folding and molecular design of proteins, and exercise a significant impact to the investigation on the origin of life.
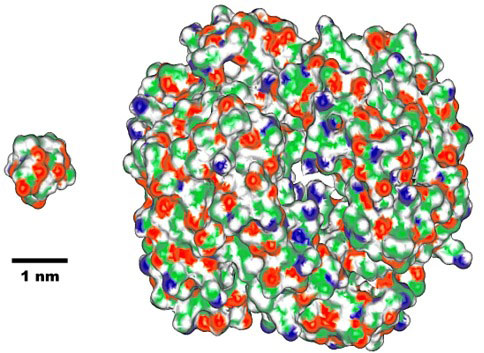 |
|
Fig. 2. Comparison of molecular size between chignolin (left, with 10 amino acid residues) and human hemoglobin, one of representative protein (right, with 574 residues). (1 nm = 1 / 1,000,000,000 m) |
The result of this work was published in the Aug. 10, 2004 issue of a U.S. journal Structure.[1]
Protein is one of most important materials supporting life phenomena, and it has been known that for the expression of its function, the formation of specific 3D structure is playing a key role. Chemically speaking, a protein is a linear heteropolymer. It is one of unsolved problems of molecular biology through what molecular mechanism and how the chain-like molecule folds into specific 3D structure, called “folding problem”. Besides, the chemical evolution of protein, that is, how and through what process, complicated and exquisite 3D structure of protein, as we see today, has been constructed, constitutes an appealing issue related to the origin of life. While both problems are far from total clarification, studies are in steady progress and many scientists are attracted to understanding of events occurring or having occurred in the initial stage. Since these studies are related not only to basic research, but also to technological foundation for identifying the process and creating new proteins, they are drawing attentions in industrial areas such as pharmaceutical, agricultural and fishery.
The IBRF-AIST has been actively engaged in the study on protein folding from earlier, and achieved a number of internationally recognized outcomes. Dr. Shinya Honda and his colleagues in the IBRF came up with an idea that there were shared molecular mechanisms between the initial stage of folding and that in chemical evolution of protein, and proposed so-called “autonomous element hypothesis” as an extension of this concept. According to this hypothesis, a protein molecule is a hierarchical structure composed of units of much smaller size than generally recognized. On the basis of this hypothesis, it has been attempted to verify its validity through the design and synthesis of protein of small size having been considered hardly realizable up to now.
It has been known that in some protein, specific sites of chain polymer called “nucleus” begins to form 3D structure prior to other sites in the earlier phase of folding. If there is any correlation between the initial process of folding and the earlier phase of molecular evolution, it may be expected that the “nucleus” structure is conserved better than other parts. Segments of 3D structure similar to that in “nucleus” of certain protein were extracted from a protein structure database. Since no software for this operation was available at the time of starting the work, an original algorithm was created to develop a special purpose computer program. Through the statistical analysis for a number of extracted segments, it was found that there was a significant bias in amino acid sequence of these segments, and that the bias reflected the stability of the partial structures concerned semi-quantitatively. With such an original computer analysis, some novel amino acid sequences were designed in expectation of forming stable structures, and named “chignolin”.
After having confirmed that synthesized chignolins were well soluble in aqueous solution, its 3D structure was analyzed through the nuclear magnetic resonance (NMR) method. The structural data obtained presented very little variation and the atomic coordinates were determined with the same accuracy as that in native proteins. The detailed analysis of denaturation/renaturation process of chignolin revealed cooperative response of entire molecule and two-state structural transition. The transition occurred broadly over a wide range of temperatures. About 20 % of chignolin molecules in equilibrium state hold 3D structures even at temperatures around 100 °C. It is very interesting that this small peptide, chignolin can persist stably under higher temperatures in view of “deep sea hydrothermal vents” hypothesis of origin of life.
Currently, the structural coordinate of chignolin has been registered in the Protein Data Bank (PDB), an international public database under collaborative operation of Japan, the United States and Europe (accession no. 1UAO). Among proteins registered in the PDB with verified both specific 3D structure and cooperative structural transition, chignolin is of smallest size.
[1] Honda, S., Yamasaki, K.. Sawada, Y. and Morii, H. (2004) 10-residue folded peptide designed by segment statistics. Structure 12(8) 1507-1518.